Optimal Local Alignment Scores for 10,000 Pairs of Random DNA Sequences
10,000 pairs of random DNA sequences are generated. Each DNA sequence is 50 nucleotides in length and has a GC content of 50%. The optimal local pairwise alignment is computed for the 10,000 pairs of sequences, so that 10,000 optimal local alignment scores are obtained.
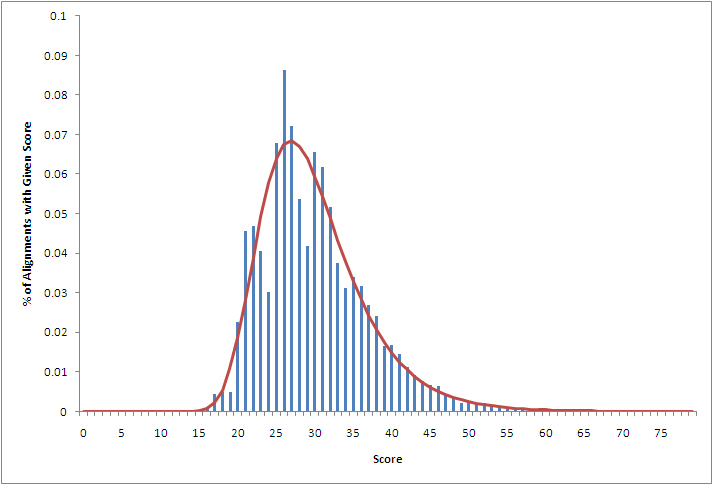
| Score | Actual # of Alignments with Given Score | Actual % of Alignemnts with Given Score | Estimated % of Alignments with Given Score |
| 0 | 0 | 0 | 1.85E-63 |
| 1 | 0 | 0 | 1.20E-52 |
| 2 | 0 | 0 | 1.10E-43 |
| 3 | 0 | 0 | 2.94E-36 |
| 4 | 0 | 0 | 4.18E-30 |
| 5 | 0 | 0 | 5.20E-25 |
| 6 | 0 | 0 | 8.57E-21 |
| 7 | 0 | 0 | 2.63E-17 |
| 8 | 0 | 0 | 2.00E-14 |
| 9 | 0 | 0 | 4.79E-12 |
| 10 | 0 | 0 | 4.38E-10 |
| 11 | 0 | 0 | 1.81E-08 |
| 12 | 0 | 0 | 3.83E-07 |
| 13 | 0 | 0 | 4.69E-06 |
| 14 | 0 | 0 | 3.64E-05 |
| 15 | 2 | 2.00E-04 | 1.93E-04 |
| 16 | 12 | 0.0012 | 7.49E-04 |
| 17 | 43 | 0.0043 | 0.00223283 |
| 18 | 49 | 0.0049 | 0.005360715 |
| 19 | 49 | 0.0049 | 0.010747809 |
| 20 | 225 | 0.0225 | 0.018553753 |
| 21 | 455 | 0.0455 | 0.028287084 |
| 22 | 470 | 0.047 | 0.038899761 |
| 23 | 405 | 0.0405 | 0.049103393 |
| 24 | 301 | 0.0301 | 0.057729104 |
| 25 | 679 | 0.0679 | 0.063979127 |
| 26 | 863 | 0.0863 | 0.067513994 |
| 27 | 722 | 0.0722 | 0.068402894 |
| 28 | 538 | 0.0538 | 0.067000843 |
| 29 | 418 | 0.0418 | 0.063811937 |
| 30 | 656 | 0.0656 | 0.059375523 |
| 31 | 618 | 0.0618 | 0.054189337 |
| 32 | 518 | 0.0518 | 0.048668355 |
| 33 | 375 | 0.0375 | 0.043131003 |
| 34 | 311 | 0.0311 | 0.037802909 |
| 35 | 340 | 0.034 | 0.032829902 |
| 36 | 317 | 0.0317 | 0.028294374 |
| 37 | 270 | 0.027 | 0.024231438 |
| 38 | 241 | 0.0241 | 0.020643051 |
| 39 | 165 | 0.0165 | 0.017509425 |
| 40 | 169 | 0.0169 | 0.014797731 |
| 41 | 144 | 0.0144 | 0.012468405 |
| 42 | 113 | 0.0113 | 0.010479514 |
| 43 | 91 | 0.0091 | 0.008789619 |
| 44 | 78 | 0.0078 | 0.007359539 |
| 45 | 66 | 0.0066 | 0.006153325 |
| 46 | 65 | 0.0065 | 0.0051387 |
| 47 | 44 | 0.0044 | 0.004287147 |
| 48 | 36 | 0.0036 | 0.00357378 |
| 49 | 21 | 0.0021 | 0.002977091 |
| 50 | 27 | 0.0027 | 0.002478627 |
| 51 | 22 | 0.0022 | 0.002062656 |
| 52 | 20 | 0.002 | 0.001715826 |
| 53 | 14 | 0.0014 | 0.001426854 |
| 54 | 10 | 0.001 | 0.001186231 |
| 55 | 9 | 9.00E-04 | 9.86E-04 |
| 56 | 6 | 6.00E-04 | 8.19E-04 |
| 57 | 4 | 4.00E-04 | 6.81E-04 |
| 58 | 4 | 4.00E-04 | 5.66E-04 |
| 59 | 2 | 2.00E-04 | 4.70E-04 |
| 60 | 5 | 5.00E-04 | 3.90E-04 |
| 61 | 1 | 1.00E-04 | 3.24E-04 |
| 62 | 1 | 1.00E-04 | 2.69E-04 |
| 63 | 2 | 2.00E-04 | 2.24E-04 |
| 64 | 2 | 2.00E-04 | 1.86E-04 |
| 65 | 0 | 0 | 1.54E-04 |
| 66 | 1 | 1.00E-04 | 1.28E-04 |
| 67 | 0 | 0 | 1.06E-04 |
| 68 | 0 | 0 | 8.83E-05 |
| 69 | 0 | 0 | 7.33E-05 |
| 70 | 0 | 0 | 6.09E-05 |
| 71 | 1 | 1.00E-04 | 5.05E-05 |
| 72 | 0 | 0 | 4.20E-05 |
| 73 | 0 | 0 | 3.48E-05 |
| 74 | 0 | 0 | 2.89E-05 |
| 75 | 0 | 0 | 2.40E-05 |
| 76 | 0 | 0 | 1.99E-05 |
| 77 | 0 | 0 | 1.66E-05 |
| 78 | 0 | 0 | 1.37E-05 |
| 79 | 0 | 0 | 1.14E-05 |