How to turn in this Project
You are required to turn in both a hardcopy and a softcopy. If working as a team, the team should submit a single hardcopy and a single softcopy (to either team member's account). Please make sure to keep a copy of your work, on your computer, in your private directory (or, to play it safe, both).Hardcopy Submission
Your hardcopy packet should consist of:- The cover page;
- Your
RNA_Hybridization.javafile from Task 1.
Softcopy Submission
You should submit your final version of your
RNA_Structure directory to the
drop/project7 directory in your account
on the CS server.
After years of toiling at a large pharmaceutical company, you decide that you are ready for a change. Following conversations with a few professional contacts, you learn of a non-profit organization that, in collaboration with a local hospital, has recently embarked on a project studying a particular form of cancer. The organization and hospital have already collected a rich set of clinical data and now they are hoping to integrate their clinical data with genomic information. Presently, they are looking for someone to lead their efforts to characterize RNA regulators that hybridize with mRNAs from a family of oncogenes that they have identified. The position intrigues you because the work is meaningful, though, admittedly, not as meaningful as your CS313 projects from oh so long ago that you look back on nostalgically. The organization is delighted to have someone of your caliber apply for the position and they hire you immediately.
Background
While chromosomal DNA is generally double stranded, RNA is often single stranded. Many RNA genes act as regulators of other genes by basepair binding to target mRNAs, affecting the translation and stability of the messages. For example, let-7 is a microRNA that can interact with and regulate RAS mRNAs. RAS proteins are proto-oncogenes that regulate cell growth and differentiation. Mutations in RAS oncogenes are present in approximately a quarter of all human cancers. let-7 can basepair bind to RAS mRNAs and inhibit their translation. Loss or reduction of let-7 leads to RAS overexpression and can promote cell growth contributing to tumorigenesis. Thus, let-7 acts as a tumor suppressor. The figure below illustrates the putative interaction between the 22 nucleotide long let-7 microRNA and part of a (reversed) RAS mRNA:
let-7 5' UGAGGUA-GUAGGUUGUAUAGUU 3'
||||||| | |||||| |||
RAS mRNA 3' GCUCCGUGCUCUUAGCGAACCAAG 5'
In the example above, note that the RAS mRNA sequence is reversed, i.e., the sequence is written from its 3' end to its 5' end. One of the sequences in the interaction is reversed because two hybridizing nucleic acid molecules interact in an anti-parallel fashion.
Task 1: RNA Hybridization
In this project, your goal is to read in two RNA sequences from files and implement an algorithm that computes the most energetically favorable interaction between the two RNAs. Here, we are not concerned with intramolecular interactions, i.e., basepairing within a given RNA molecule. We are only concerned with intermolecular interactions, i.e., basepairing between nucleotides of two different RNA molecules.
In class, when we computed the most energetically favorable structure of a single
RNA molecule, we considered a variety of structural elements including hairpin loops,
stacking basepairs, bulge loops, interior loops, and bifurcations. Here, since
we are only concerned with intermolecular interactions, we need only consider
a subset of the abovementioned structural elements, namely stacking basepairs,
bulge loops, and interior loops. Given two RNA molecules, S and T,
with lengths n and m, respectively, the most energetically
favorable hybridization of the two sequences can be described by the following
recurrence:
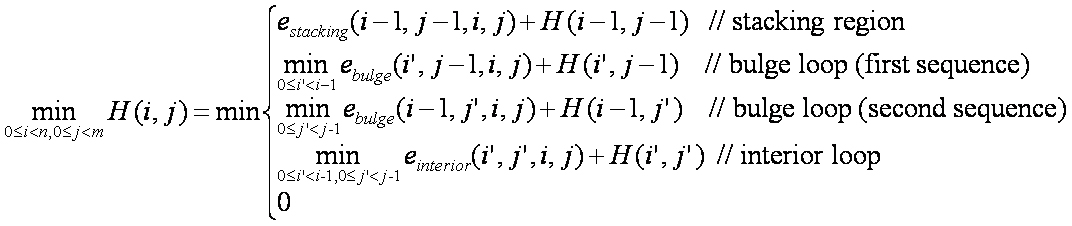
In the recurrence above, estacking, ebulge, and einterior refer to the energies of stacking regions, bulge loops, and interior loops, respectively. The variable i represents an index in sequence S, the variable j represents an index in sequence T, and H(i, j) represents the minimum energy of hybridization between subsequence S(0...i) and subsequence T(0...j).
The above recurrence enables computation of the optimal (minimum) energy
of hybridization of two RNA sequences. If we want to determine the set of
hybridizing basepairs that yield the optimal energy of hybridization, we
can do so via backtracking. In the let-7 example above,
there are three lines, which we will denote with the names upper,
hybridization, and lower. upper refers to the first
sequence (including any gaps) participating in the hybridization, e.g.,
UGAGGUA-GUAGGUUGUAUAGUU. hybridization refers to the
sequence of vertical bars that indicates interacting basepairs from the two
sequences, e.g., ||||||| | |||||| |||. lower refers
to the second sequence (including any gaps) participating in the hybridization,
e.g., GCUCCGUGCUCUUAGCGAACCAAG. The pseudocode below suggests
one approach for computing the three lines that represent the hybridization
with optimal energy.

In the pseudocode above, spaces(x) refers to a function that generates a String of x space characters, and gaps(x) refers to a function that generates a String of x gap characters, '-'.
To begin, download the RNA_Structure folder from the
download folder on the CS server.
This folder contains a file RNA_Energy.java
that may be useful as a point of reference as well as a class
Energies with several useful helper methods. The
contract for the Energies class can be found here.
Your goal is to design and implement a class,
RNA_Hybridization, that reads in RNA sequences from two
FASTA files specified by command line arguments, and computes the optimal
hybridization of the two RNAs. Your program should output both the energy
of the optimal hybridization as well as a text representation
of the basepairs participating in the optimal hybridization. For example,
executing your program might result in the following output:
java RNA_Hybridization sequences/RNA_2a.txt sequences/RNA_2b.txt
Hybridization with lowest energy:
AGGCAUUCAGAAUG
||| ||| |||
ACCGCAAG-CUUC
Optimal hybridization energy: -10.8
With the code available from the download folder on the
CS server, we have provided you with a
sample solution, TEST_RNA_Hybridization, which you can execute
and compare with your own solution (actually, the sample solution outputs
more information than necessary to aid you in debugging).